Bioinformatics Services We Provide Here:
At Genetic Codon, we provide comprehensive bioinformatics services tailored to support academic researchers, healthcare professionals, and biotech companies. Our team uses state-of-the-art methodologies and tools to help you turn complex biological data into meaningful insights. Explore our services and take your research to the next level.
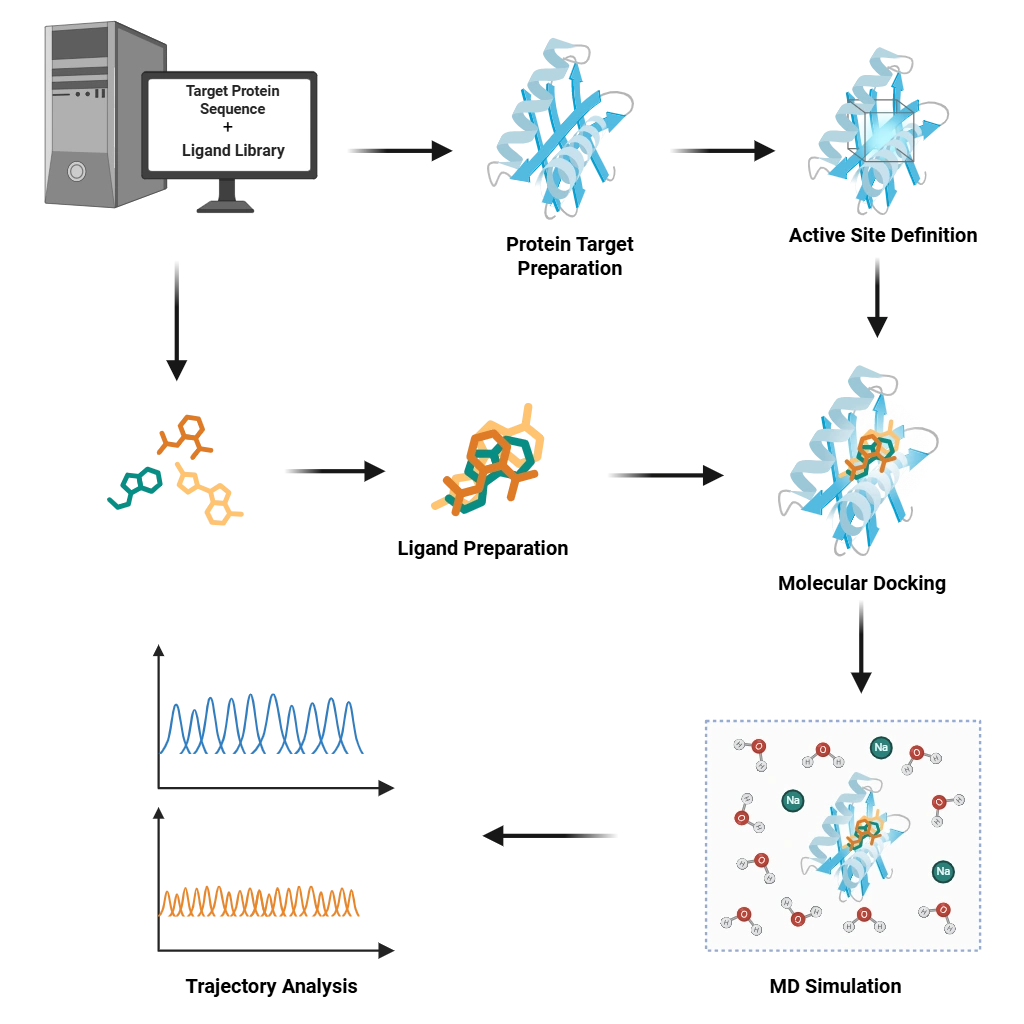
Structural Biology Services
We offer computational modeling, molecular docking, and dynamics simulations to understand biomolecular structures and interactions. Techniques: Homology modeling, molecular dynamics (GROMACS, AMBER), docking (AutoDock, HADDOCK).
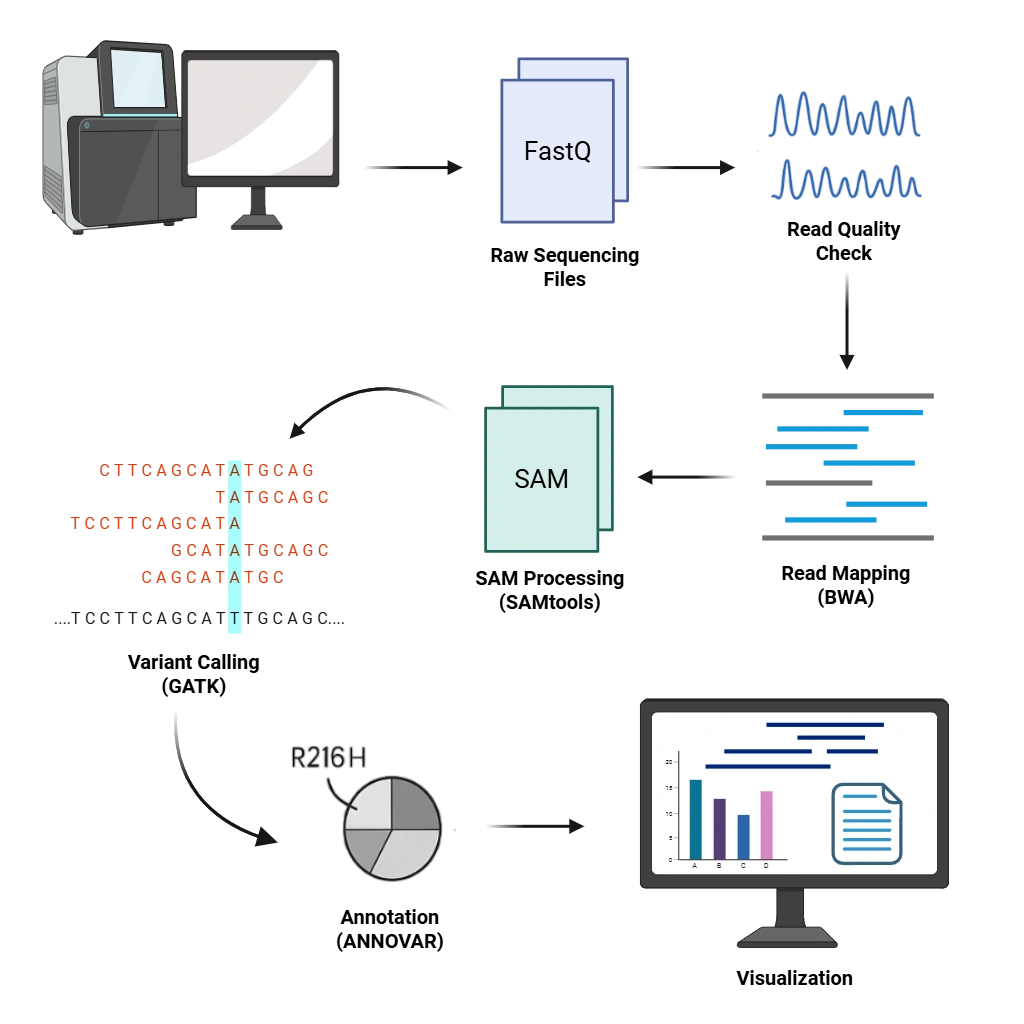
Whole Genome Sequencing (WGS) Data Analysis
From raw sequencing files to meaningful biological insights, we process and analyze full genomes using industry-standard pipelines (GATK, BWA, SAMtools) for variant detection, annotation, and visualization.

Whole Exome Sequencing (WES) Data Analysis
Targeted analysis of coding regions to identify pathogenic mutations. Includes alignment, variant calling, annotation, and downstream interpretation using cutting-edge tools like VarScan and ANNOVAR.
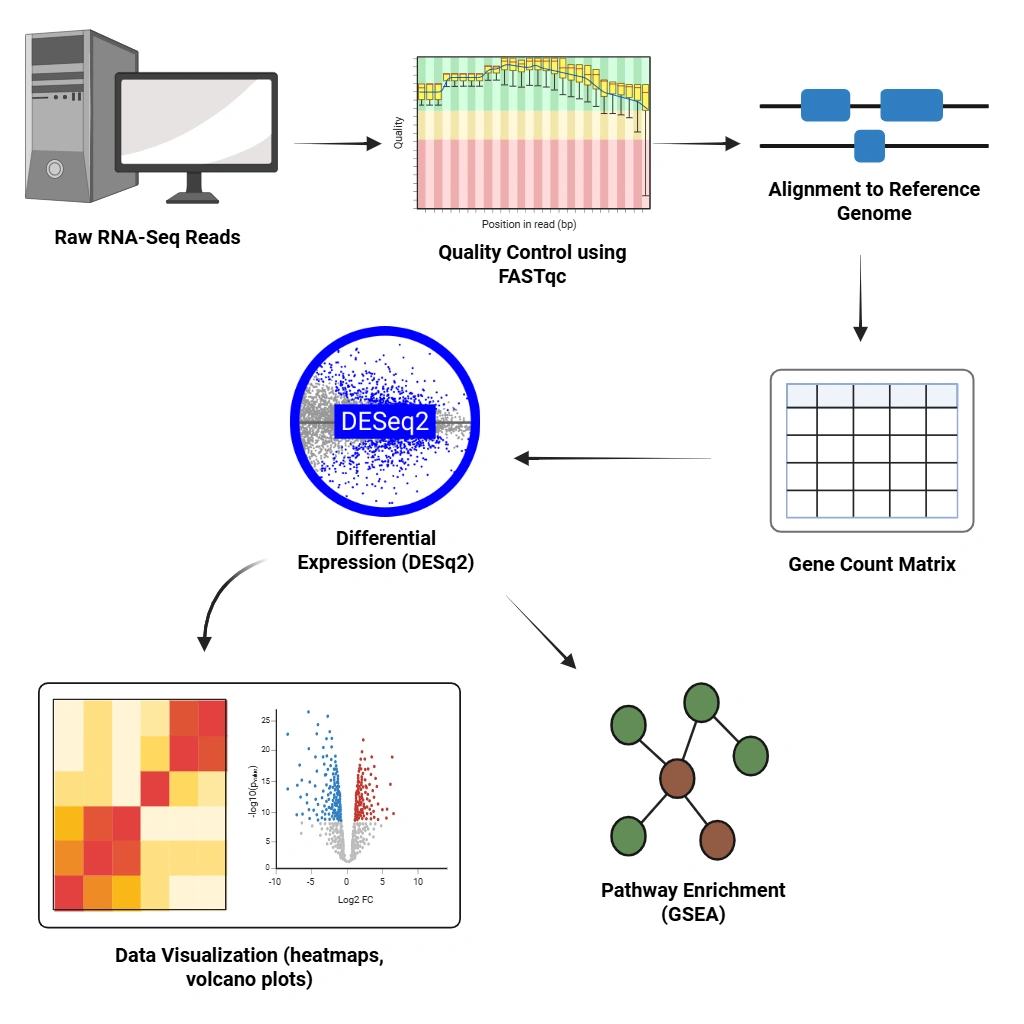
Bulk Transcriptome Data Analysis (RNA-Seq)
Get deep insights into gene expression dynamics. We perform differential expression analysis (DESeq2, EdgeR), pathway enrichment (GSEA), and visualization (heatmaps, volcano plots).
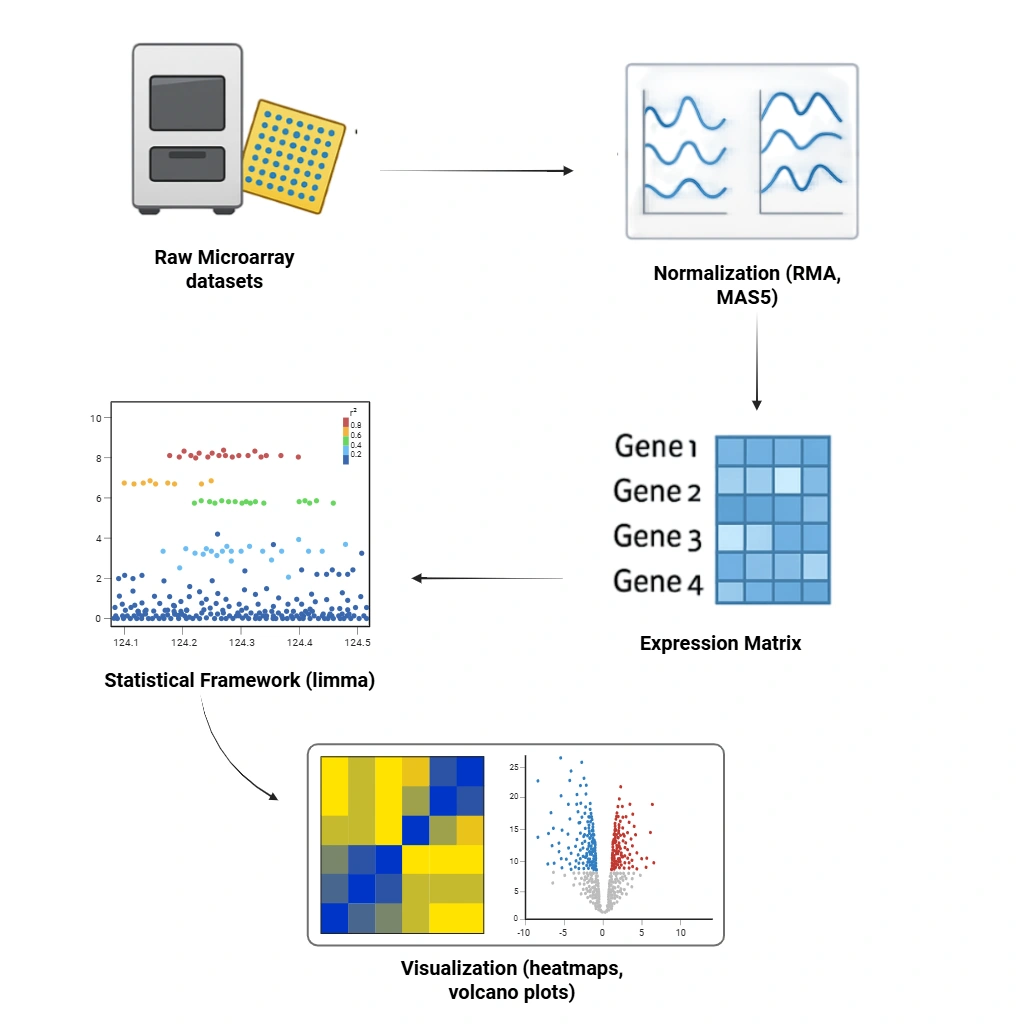
Microarray RNA Expression Data Analysis
We analyze microarray datasets for differential gene expression using normalization techniques
(RMA, MAS5) and statistical frameworks
(limma).
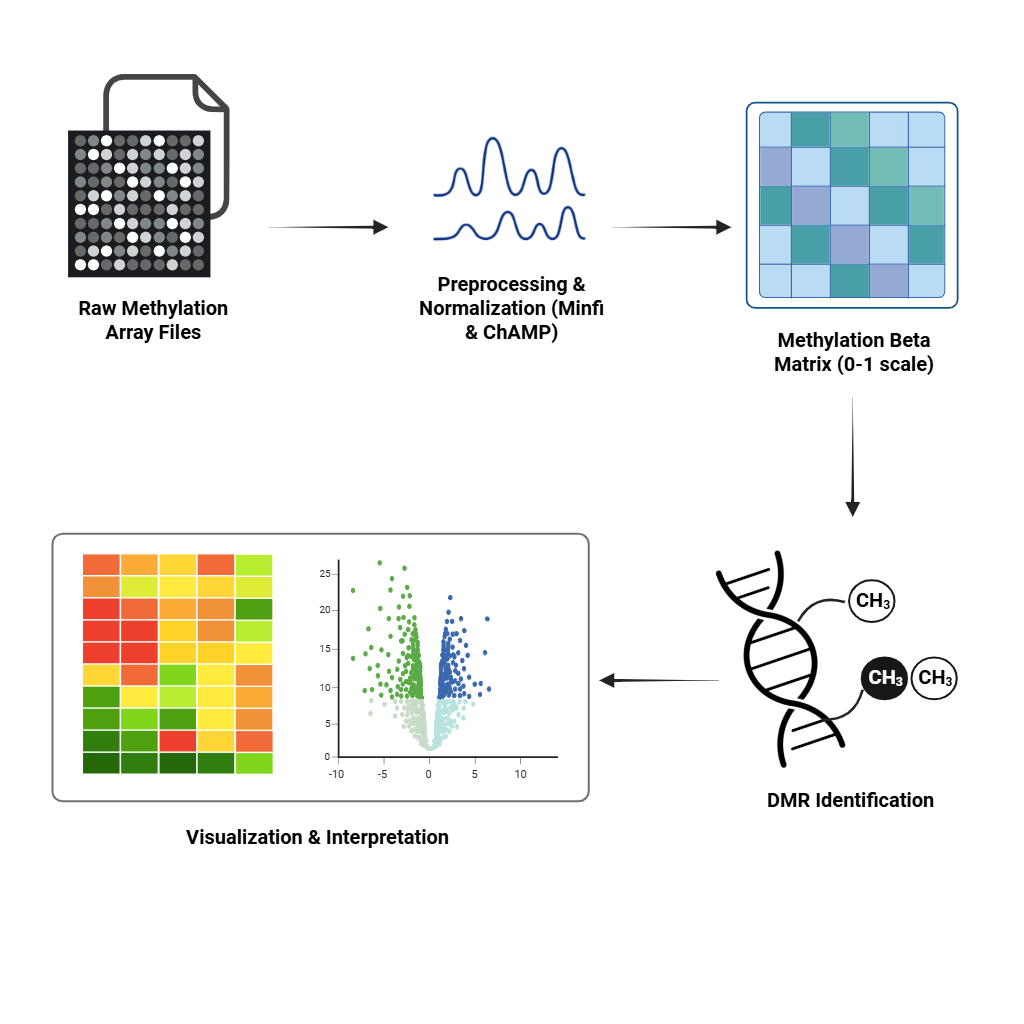
Microarray DNA Methylation Data Analysis
Quantify and interpret methylation changes using tools like Minfi and ChAMP. From preprocessing to identification of differentially methylated regions (DMRs).
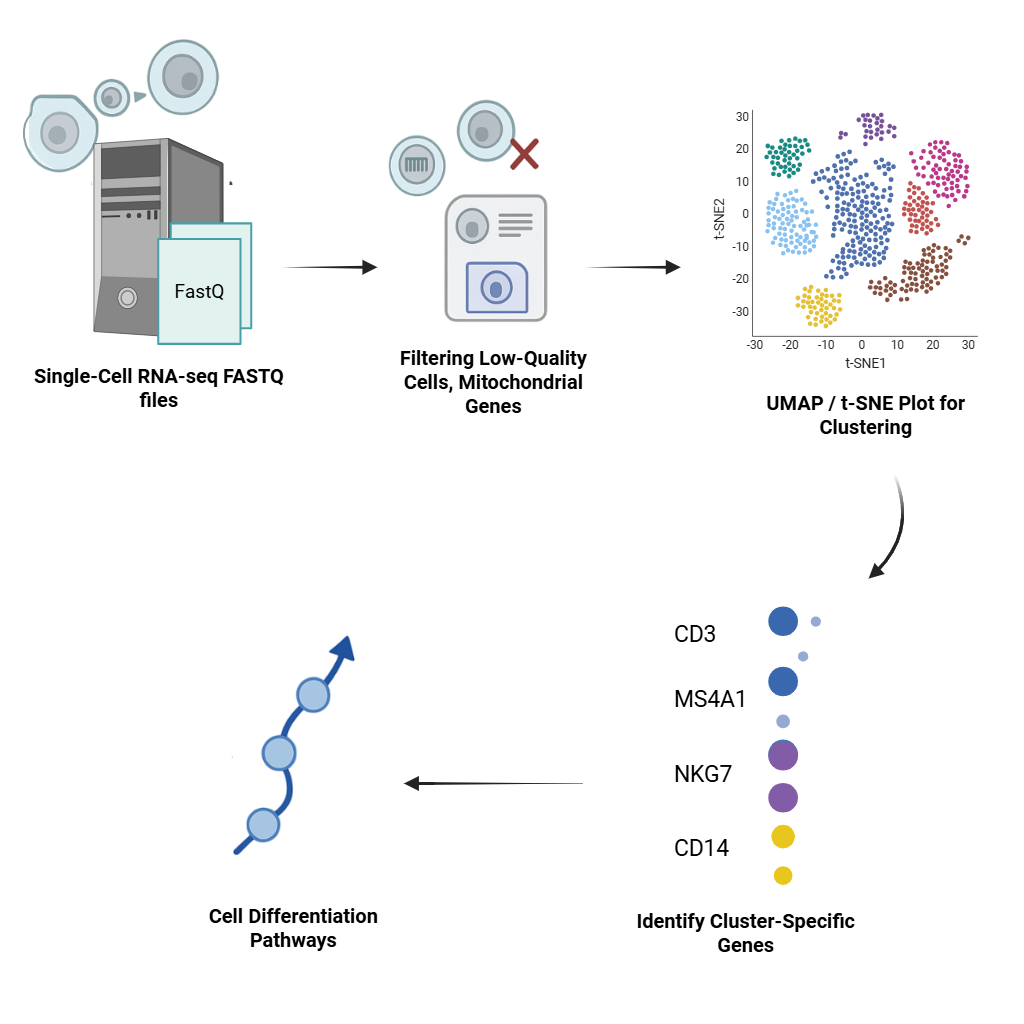
Single Cell Data Analysis
Unlock the complexity of cellular heterogeneity. We use Seurat, Scanpy, and Harmony for clustering, trajectory inference, and marker gene identification at single-cell resolution
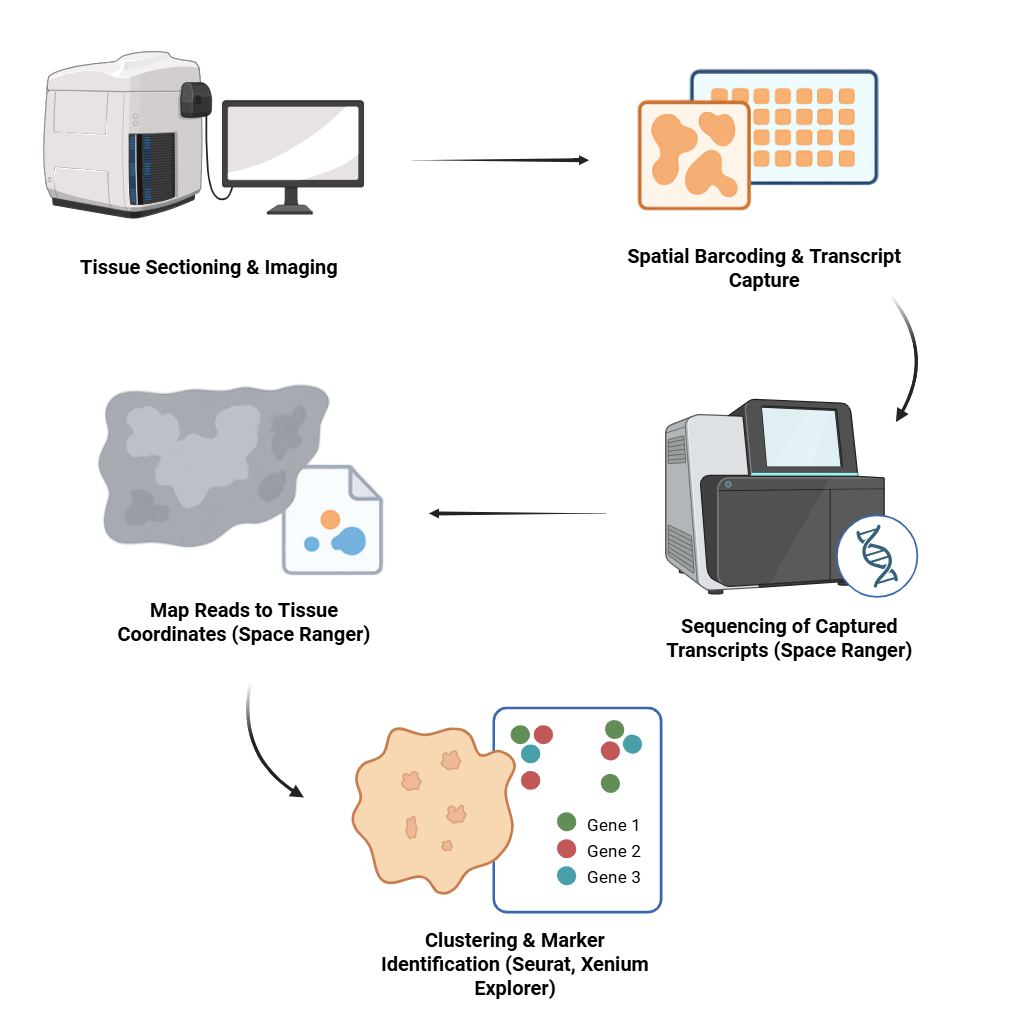
Spatial Data Analysis (Visium HD/Xenium)
We offer complete spatial gene expression analysis. Tools used include Space Ranger, Seurat v4, and Xenium Explorer.
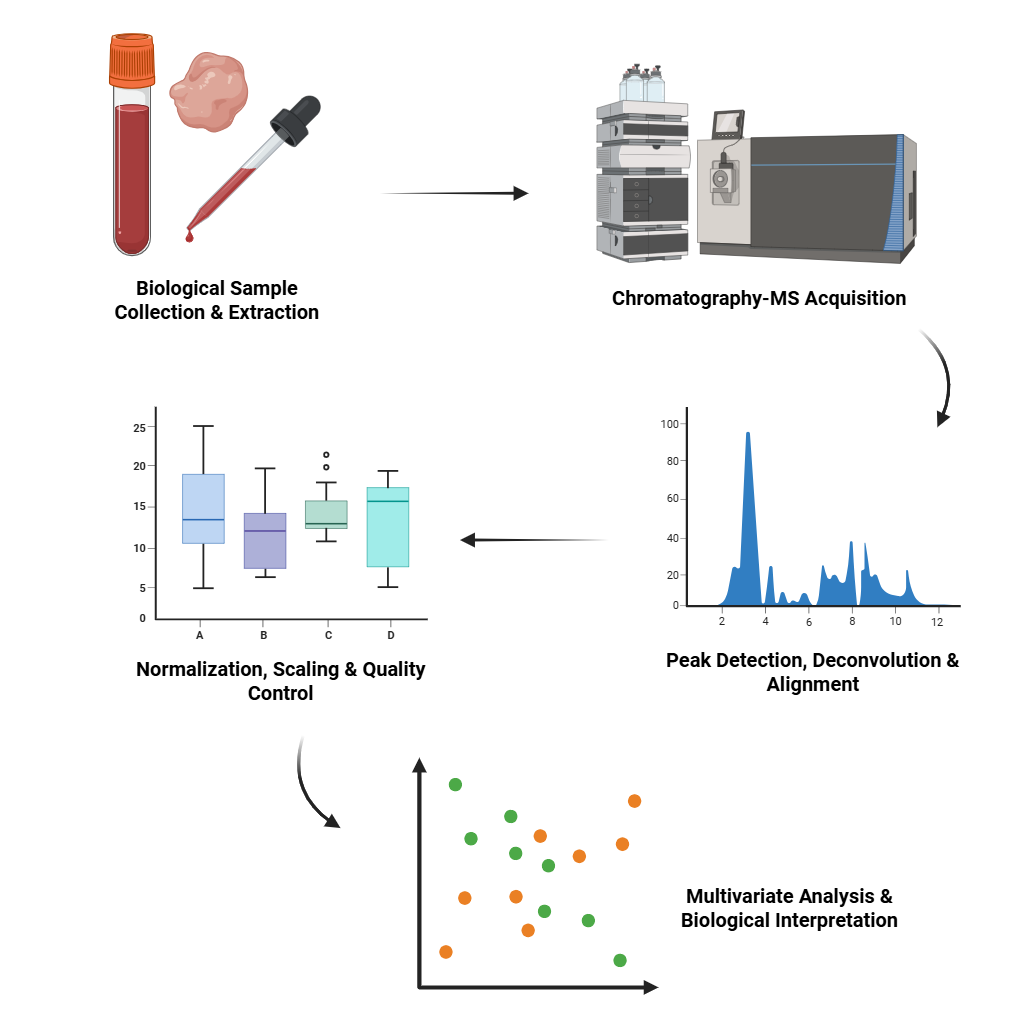
Metabolomics Data Analysis
Analyze metabolite profiles from LC-MS/GC-MS platforms. We support preprocessing, peak identification, normalization, and multivariate analysis (PCA, PLS-DA).
Proteomics
Data Analysis
From raw MS/MS data to functional interpretation, we offer protein identification, quantification, and pathway analysis using MaxQuant, Perseus, and IPA.
Meta-analysis and Systematic Reviews
Consolidate findings across multiple studies. We use PRISMA guidelines and statistical methods (meta, metafor packages in R) for robust meta-analyses in biomedical research.
Copy Number Variation (CNV) Data Analysis
Detection and interpretation of CNVs from WGS/WES data using tools like CNVkit and XHMM, supporting disease association and functional studies.
Metagenomics Data Analysis
Explore microbial communities through 16S rRNA or shotgun metagenomics sequencing. We provide taxonomy assignment (QIIME2, Kraken2) and functional profiling (HUMAnN2).
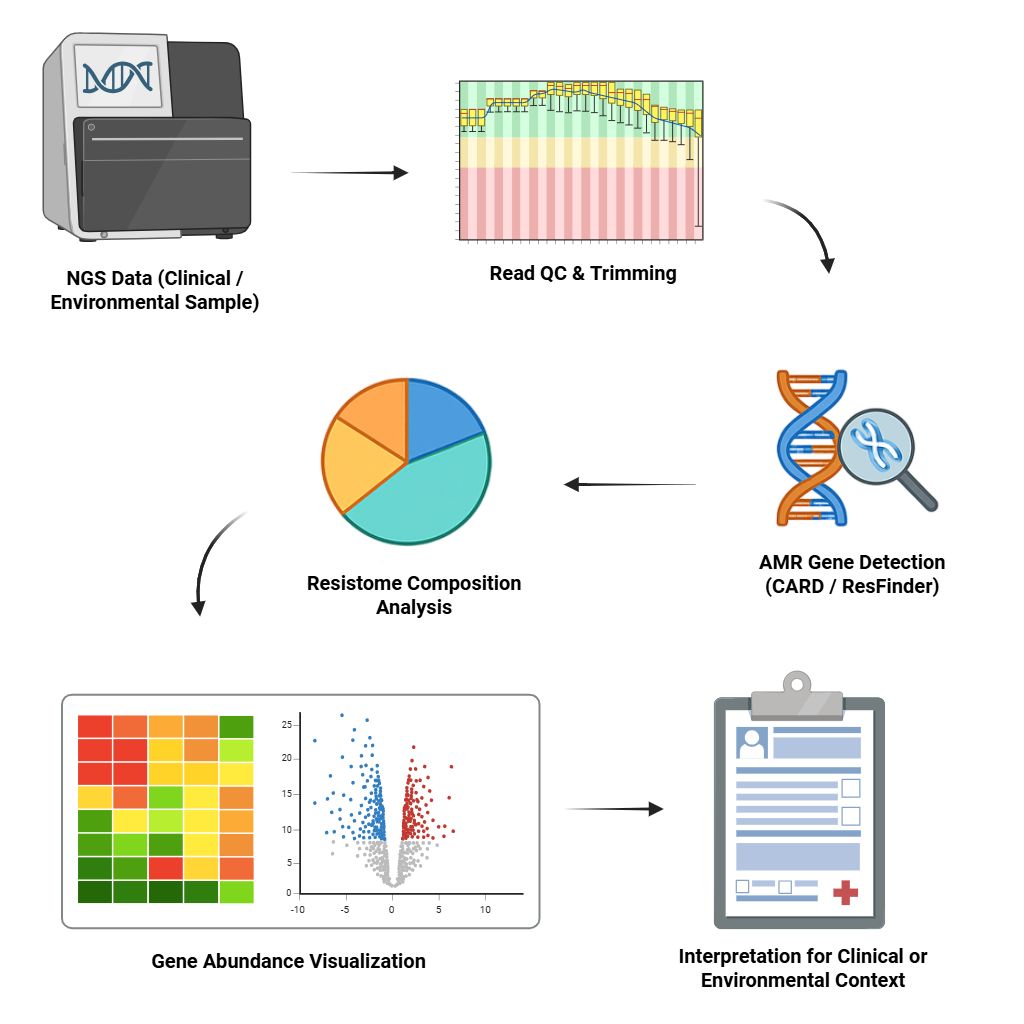
Antimicrobial Resistance (AMR)
Identify AMR genes using databases (CARD, ResFinder), interpret the resistome from NGS datasets for clinical or environmental studies.
For any inquiries, please email
Interested in availing our services or learning more?
info@geneticocodon.com

- Head Office
- Pakistan
- info@geneticcodon.com
- +92 333 122 4495
- Quick Links
- Resources
